Quick costum bioanalyzer plots using R
The default plots made by the bioanalyzer are, in my opinion, not amazing. For one, the lines are really thin. Furthermore, it would be nice to draw two traces is the same plot, to compare different samples. Luckily, the bioanalyzer software lets you export the raw data. This short post will show how to quickly get bioanalyzer trace data into R and ready for plotting.
On the way we will make use of the tidyverse packages.
library(tidyverse)Importing one file into R
You can get the raw data out of the bioanalyzer by going File > Export. Somewhat depending on the options you select next, it will spit out many different files:
list.files(path = "./20210106data")## [1] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52.xml"
## [2] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_GEL.bmp"
## [3] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_GEL_Ladder.bmp"
## [4] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_GEL_Sample1.bmp"
## [5] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_GEL_Sample2.bmp"
## [6] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_GEL_Sample3.bmp"
## [7] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_GEL_Sample4.bmp"
## [8] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_GEL_Sample5.bmp"
## [9] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_GEL_Sample6.bmp"
## [10] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Ladder.csv"
## [11] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Results.csv"
## [12] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample1.csv"
## [13] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample2.csv"
## [14] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample3.csv"
## [15] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample4.csv"
## [16] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample5.csv"
## [17] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample6.csv"Annoyingly, the raw data for each well of the bioanalyzer chip is found in a separate csv file. Manually pasting these together in excel is too much clicking, and more importantly error prone. Depending on your location, excel may see the comma as the thousand separator, which makes converting the csv file into excel columns with Data > Text to Columns go wrong for some rows (specifically those with a round number for time and high fluorescence, like 40,123). Importing the csv files into R will not run into this problem.
I generally start out by creating an R project in the folder containing the bioanalyzer data. Among other things, this avoids having to set the working directory.
The next step is to separate the csv files containing the raw data from the rest. These files always end in “Sample” + the well number + “.csv” (at least for RNA nano and pico chips). We can use this pattern to isolate them, by creating a regular expression and feeding this pattern to list.files(). We are not interested in what comes before Sample, so we can preface Sample with the * symbol. This means zero or more of anything can occur. Sample is always followed by one or more digits. We tell the function to look for a digit using \\d and use + to allow for one or more them. Finally, all these files should end in .csv. The dollar sign signifies that the string should end there.
# Ignore the path = , this is only here because of how the website is structured.
# If you have made an r project inside the folder containing the bioanalyzer
# data you won't need it.
samples <- list.files(path = "./20210106data", pattern = "*Sample\\d+.csv$")
samples## [1] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample1.csv"
## [2] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample2.csv"
## [3] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample3.csv"
## [4] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample4.csv"
## [5] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample5.csv"
## [6] "2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample6.csv"It worked!
Let’s look at what happens when we import one of these files.
# Again, the path = , and paste0 shenanigans is only needed for the website.
# In a normal situation just read_csv(samples[1]) would be enough.
path <- "./20210106data/"
sample1 <- read_csv(paste0(path, samples[1]))| Data File Name | 2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52.xad |
|---|---|
| Data File Path | C:Files (x86)\2100 bioanalyzer\2100 expert\2020-05-25 |
| Date Created | Monday, May 25, 2020 9:42:52 AM |
| Date Last Modified | Monday, May 25, 2020 10:06:42 AM |
| Version Created | B.02.10.SI764 |
| Version Last Modified | B.02.10.SI764 |
| Assay Name | Eukaryote Total RNA Nano Series II |
| Assay Path | C:Files (x86)\2100 bioanalyzer\2100 expert |
| Assay Title | Eukaryote Total RNA Nano |
| Assay Version | 2.6 |
| Number of Samples Run | 12 |
| Sample Name | 0 ctrk |
| Number of Events | 1060 |
| Time | Value |
| 17 | 1.657798E-02 |
| 17.05 | 2.474964E-02 |
The first few rows are all metadata we are not really interested in. We only need the Time and Value information. Therefore, we can skip the first 17 rows (including some empty rows).
sample1 <- read_csv(paste0(path, samples[1]),
skip = 17)## Warning: 2 parsing failures.
## row col expected actual file
## 1061 Time a double Alignment './20210106data/2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample1.csv'
## 1061 Value a double On './20210106data/2100 expert_Eukaryote Total RNA Nano_DE13805377_2020-05-25_11-42-52_Sample1.csv'But now we get 2 parsing failures. That is because, all the way at the bottom of the file, some text (“alignment, on”) appears where only numbers are expected, which results in NA values in the final row of our table.
| Time | Value |
|---|---|
| 69.70 | 0.7465172 |
| 69.75 | 0.7465172 |
| 69.80 | 0.7465172 |
| 69.85 | 0.7465172 |
| 69.90 | 0.7465172 |
| 69.95 | 0.7465172 |
| NA | NA |
To prevent this we can tell read_csv to not read this far into the file.
sample1 <- read_csv(paste0(path, samples[1]),
skip = 17,
n_max = 1060)No failures this time!
Importing multiple files into r
We have successfully imported the information of one well into r. But there are 11 or 12 wells on a chip. One option is to simply copy paste the code 12 times and change the numbers each time. But this leads to a lot of lines of code and increases the risk of errors.
sample1 <- read_csv(paste0(path, samples[1]),
skip = 17,
n_max = 1060)
sample2 <- read_csv(paste0(path, samples[2]),
skip = 17,
n_max = 1060)
# O no, a typo
sample2 <- read_csv(paste0(path, samples[3]),
skip = 17,
n_max = 1060)
# Etc. Instead, we can use a single for loop to import all the data files. In this loop we also save them into one big data table. We will use mutate to make a new column that contains the well number each measurement originates from. str_extract is used to extract this information from the file name.
# Create an empty table to save the data in.
all_data <- tibble()
for(i in samples){
# Import one by one
data <- read_csv(paste0(path, i),
skip = 17,
n_max = 1060)
# Add the well number information
data <- mutate(data, sample_nr = str_extract(i, pattern = "Sample\\d+"))
# Store in the big table
all_data <- rbind(all_data, data)
}| Time | Value | sample_nr |
|---|---|---|
| 17.00 | 0.0165780 | Sample1 |
| 17.05 | 0.0247496 | Sample1 |
| 17.10 | 0.0391493 | Sample1 |
| 17.15 | 0.0626396 | Sample1 |
| 17.20 | 0.0790722 | Sample1 |
| 17.25 | 0.0928879 | Sample1 |
| 17.30 | 0.0869223 | Sample1 |
| 17.35 | 0.0712983 | Sample1 |
It would be even more convenient if the sample_nr column only contained, well … , numbers.
Again str_extract can help use remove all the unwanted symbols. Even though we are only left with numbers, we still need to convert it to a numeric, because a string will stay a string until told otherwise.
all_data <- all_data %>%
mutate(sample_nr = as.numeric(str_extract(sample_nr, pattern = "\\d+$")))| Time | Value | sample_nr |
|---|---|---|
| 17.00 | 0.0165780 | 1 |
| 17.05 | 0.0247496 | 1 |
| 17.10 | 0.0391493 | 1 |
| 17.15 | 0.0626396 | 1 |
| 17.20 | 0.0790722 | 1 |
| 17.25 | 0.0928879 | 1 |
| 17.30 | 0.0869223 | 1 |
| 17.35 | 0.0712983 | 1 |
Adding sample information
Now it is also easy to add additional information about each sample. You can simply prepare a small table in excel that contains at least a sample_nr column, and any number of additional columns with extra information about each sample.
exp_design <- read_delim("20210106_exp_design.csv", delim = ";")| sample_nr | treatment | replicate |
|---|---|---|
| 1 | CTRL | 1 |
| 2 | CTRL | 2 |
| 3 | CTRL | 3 |
| 4 | treatment | 1 |
| 5 | treatment | 2 |
| 6 | treatment | 3 |
Finally we can join these two tables together based on the sample_nr column.
all_data <- all_data %>%
left_join(exp_design, by = "sample_nr")| Time | Value | sample_nr | treatment | replicate |
|---|---|---|---|---|
| 17.00 | 0.0165780 | 1 | CTRL | 1 |
| 17.05 | 0.0247496 | 1 | CTRL | 1 |
| 17.10 | 0.0391493 | 1 | CTRL | 1 |
| 17.15 | 0.0626396 | 1 | CTRL | 1 |
| 17.20 | 0.0790722 | 1 | CTRL | 1 |
| 17.25 | 0.0928879 | 1 | CTRL | 1 |
| 17.30 | 0.0869223 | 1 | CTRL | 1 |
| 17.35 | 0.0712983 | 1 | CTRL | 1 |
Saving and/or plotting
At this point you could save the final table if you want to do further analysis in other software.
write_csv(all_data, "bioanalyzer_data.csv")Or you can make plots in r!
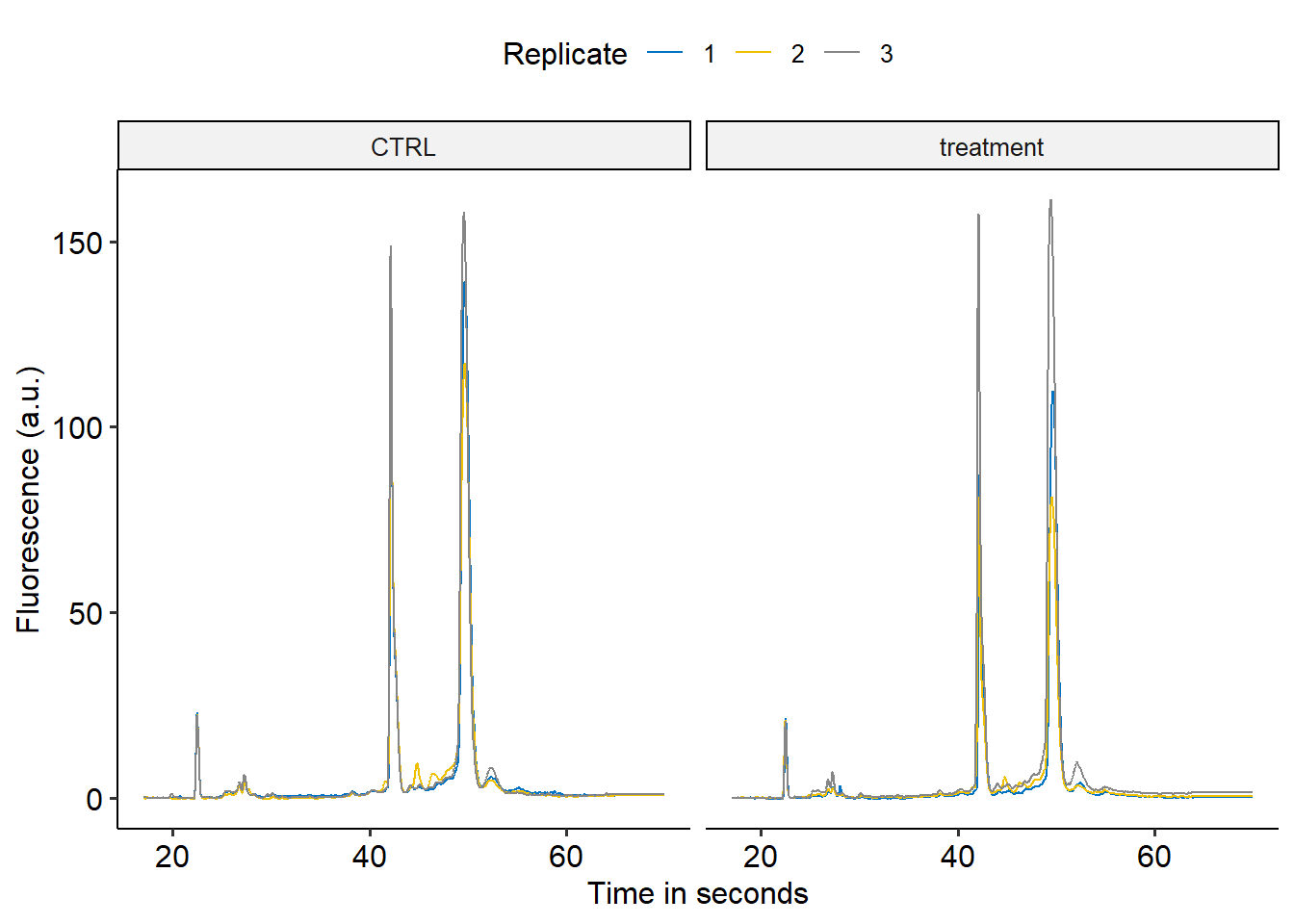
library(ggpubr)
library(ggsci)
all_data %>%
ggplot(aes(x = Time, y = Value, color = as.factor(replicate)))+
geom_line()+
facet_grid(cols = vars(treatment))+
scale_x_continuous(breaks = c(20, 40, 60))+
scale_color_jco(name = "Replicate")+
theme_pubr()+
labs(x = "Time in seconds",
y = "Fluorescence (a.u.)")